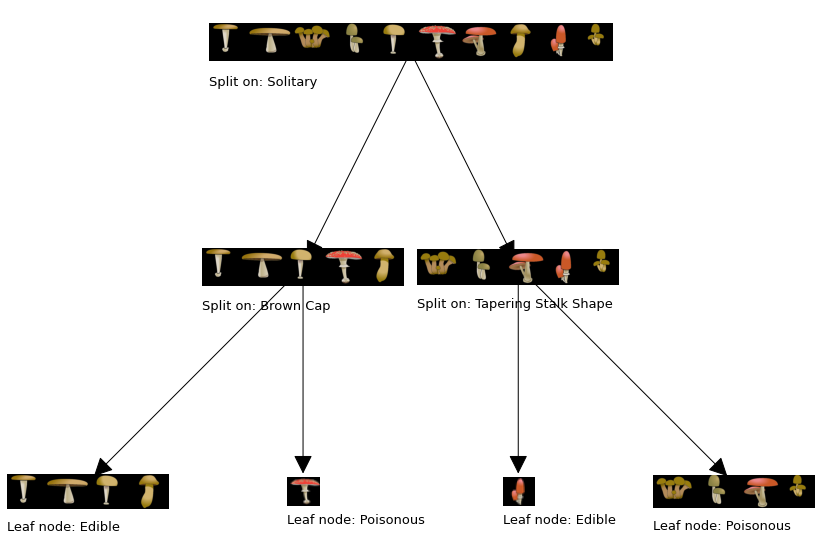Coursera
Practice Lab: Decision Trees
In this exercise, you will implement a decision tree from scratch and apply it to the task of classifying whether a mushroom is edible or poisonous.
Outline
1 - Packages
First, let’s run the cell below to import all the packages that you will need during this assignment.
- numpy is the fundamental package for working with matrices in Python.
- matplotlib is a famous library to plot graphs in Python.
utils.pycontains helper functions for this assignment. You do not need to modify code in this file.
import numpy as np
import matplotlib.pyplot as plt
from public_tests import *
from utils import *
%matplotlib inline
2 - Problem Statement
Suppose you are starting a company that grows and sells wild mushrooms.
- Since not all mushrooms are edible, you’d like to be able to tell whether a given mushroom is edible or poisonous based on it’s physical attributes
- You have some existing data that you can use for this task.
Can you use the data to help you identify which mushrooms can be sold safely?
Note: The dataset used is for illustrative purposes only. It is not meant to be a guide on identifying edible mushrooms.
3 - Dataset
You will start by loading the dataset for this task. The dataset you have collected is as follows:
| Cap Color | Stalk Shape | Solitary | Edible | |
|---|---|---|---|---|
 |
Brown | Tapering | Yes | 1 |
 |
Brown | Enlarging | Yes | 1 |
 |
Brown | Enlarging | No | 0 |
 |
Brown | Enlarging | No | 0 |
 |
Brown | Tapering | Yes | 1 |
 |
Red | Tapering | Yes | 0 |
 |
Red | Enlarging | No | 0 |
 |
Brown | Enlarging | Yes | 1 |
 |
Red | Tapering | No | 1 |
 |
Brown | Enlarging | No | 0 |
- You have 10 examples of mushrooms. For each example, you have
- Three features
- Cap Color (
BrownorRed), - Stalk Shape (
Tapering (as in \/)orEnlarging (as in /\)), and - Solitary (
YesorNo)
- Cap Color (
- Label
- Edible (
1indicating yes or0indicating poisonous)
- Edible (
- Three features
3.1 One hot encoded dataset
For ease of implementation, we have one-hot encoded the features (turned them into 0 or 1 valued features)
| Brown Cap | Tapering Stalk Shape | Solitary | Edible | |
|---|---|---|---|---|
 |
1 | 1 | 1 | 1 |
 |
1 | 0 | 1 | 1 |
 |
1 | 0 | 0 | 0 |
 |
1 | 0 | 0 | 0 |
 |
1 | 1 | 1 | 1 |
 |
0 | 1 | 1 | 0 |
 |
0 | 0 | 0 | 0 |
 |
1 | 0 | 1 | 1 |
 |
0 | 1 | 0 | 1 |
 |
1 | 0 | 0 | 0 |
Therefore,
-
X_traincontains three features for each example- Brown Color (A value of
1indicates “Brown” cap color and0indicates “Red” cap color) - Tapering Shape (A value of
1indicates “Tapering Stalk Shape” and0indicates “Enlarging” stalk shape) - Solitary (A value of
1indicates “Yes” and0indicates “No”)
- Brown Color (A value of
-
y_trainis whether the mushroom is edibley = 1indicates edibley = 0indicates poisonous
X_train = np.array([[1,1,1],[1,0,1],[1,0,0],[1,0,0],[1,1,1],[0,1,1],[0,0,0],[1,0,1],[0,1,0],[1,0,0]])
y_train = np.array([1,1,0,0,1,0,0,1,1,0])
View the variables
Let’s get more familiar with your dataset.
- A good place to start is to just print out each variable and see what it contains.
The code below prints the first few elements of X_train and the type of the variable.
print("First few elements of X_train:\n", X_train[:5])
print("Type of X_train:",type(X_train))
First few elements of X_train:
[[1 1 1]
[1 0 1]
[1 0 0]
[1 0 0]
[1 1 1]]
Type of X_train: <class 'numpy.ndarray'>
Now, let’s do the same for y_train
print("First few elements of y_train:", y_train[:5])
print("Type of y_train:",type(y_train))
First few elements of y_train: [1 1 0 0 1]
Type of y_train: <class 'numpy.ndarray'>
Check the dimensions of your variables
Another useful way to get familiar with your data is to view its dimensions.
Please print the shape of X_train and y_train and see how many training examples you have in your dataset.
print ('The shape of X_train is:', X_train.shape)
print ('The shape of y_train is: ', y_train.shape)
print ('Number of training examples (m):', len(X_train))
The shape of X_train is: (10, 3)
The shape of y_train is: (10,)
Number of training examples (m): 10
4 - Decision Tree Refresher
In this practice lab, you will build a decision tree based on the dataset provided.
-
Recall that the steps for building a decision tree are as follows:
- Start with all examples at the root node
- Calculate information gain for splitting on all possible features, and pick the one with the highest information gain
- Split dataset according to the selected feature, and create left and right branches of the tree
- Keep repeating splitting process until stopping criteria is met
-
In this lab, you’ll implement the following functions, which will let you split a node into left and right branches using the feature with the highest information gain
- Calculate the entropy at a node
- Split the dataset at a node into left and right branches based on a given feature
- Calculate the information gain from splitting on a given feature
- Choose the feature that maximizes information gain
-
We’ll then use the helper functions you’ve implemented to build a decision tree by repeating the splitting process until the stopping criteria is met
- For this lab, the stopping criteria we’ve chosen is setting a maximum depth of 2
4.1 Calculate entropy
First, you’ll write a helper function called compute_entropy that computes the entropy (measure of impurity) at a node.
- The function takes in a numpy array (
y) that indicates whether the examples in that node are edible (1) or poisonous(0)
Complete the compute_entropy() function below to:
- Compute $p_1$, which is the fraction of examples that are edible (i.e. have value =
1iny) - The entropy is then calculated as
$$H(p_1) = -p_1 \text{log}_2(p_1) - (1- p_1) \text{log}_2(1- p_1)$$
- Note
- The log is calculated with base $2$
- For implementation purposes, $0\text{log}_2(0) = 0$. That is, if
p_1 = 0orp_1 = 1, set the entropy to0 - Make sure to check that the data at a node is not empty (i.e.
len(y) != 0). Return0if it is
Exercise 1
Please complete the compute_entropy() function using the previous instructions.
If you get stuck, you can check out the hints presented after the cell below to help you with the implementation.
# UNQ_C1
# GRADED FUNCTION: compute_entropy
def compute_entropy(y):
"""
Computes the entropy for
Args:
y (ndarray): Numpy array indicating whether each example at a node is
edible (`1`) or poisonous (`0`)
Returns:
entropy (float): Entropy at that node
"""
# You need to return the following variables correctly
entropy = 0.
### START CODE HERE ###
poisson = np.sum(y)
if poisson == len(y) or poisson == 0:
entropy = 0
else:
entropy = (-poisson/len(y)) * np.log2(poisson/len(y)) - (1 - poisson/len(y)) * np.log2(1 - poisson/len(y))
### END CODE HERE ###
return entropy
Click for hints
- To calculate
p1- You can get the subset of examples in
ythat have the value1asy[y == 1] - You can use
len(y)to get the number of examples iny
- You can get the subset of examples in
- To calculate
entropy- np.log2 let’s you calculate the logarithm to base 2 for a numpy array
- If the value of
p1is 0 or 1, make sure to set the entropy to0
<details>
<summary><font size="2" color="darkblue"><b> Click for more hints</b></font></summary>
* Here's how you can structure the overall implementation for this function
```python
def compute_entropy(y):
# You need to return the following variables correctly
entropy = 0.
### START CODE HERE ###
if len(y) != 0:
# Your code here to calculate the fraction of edible examples (i.e with value = 1 in y)
p1 =
# For p1 = 0 and 1, set the entropy to 0 (to handle 0log0)
if p1 != 0 and p1 != 1:
# Your code here to calculate the entropy using the formula provided above
entropy =
else:
entropy = 0.
### END CODE HERE ###
return entropy
```
If you're still stuck, you can check the hints presented below to figure out how to calculate `p1` and `entropy`.
<details>
<summary><font size="2" color="darkblue"><b>Hint to calculate p1</b></font></summary>
    You can compute p1 as <code>p1 = len(y[y == 1]) / len(y) </code>
</details>
<details>
<summary><font size="2" color="darkblue"><b>Hint to calculate entropy</b></font></summary>
    You can compute entropy as <code>entropy = -p1 * np.log2(p1) - (1 - p1) * np.log2(1 - p1)</code>
</details>
</details>
You can check if your implementation was correct by running the following test code:
# Compute entropy at the root node (i.e. with all examples)
# Since we have 5 edible and 5 non-edible mushrooms, the entropy should be 1"
print("Entropy at root node: ", compute_entropy(y_train))
# UNIT TESTS
compute_entropy_test(compute_entropy)
Entropy at root node: 1.0
[92m All tests passed.
Expected Output:
| Entropy at root node: 1.0 |
4.2 Split dataset
Next, you’ll write a helper function called split_dataset that takes in the data at a node and a feature to split on and splits it into left and right branches. Later in the lab, you’ll implement code to calculate how good the split is.
- The function takes in the training data, the list of indices of data points at that node, along with the feature to split on.
- It splits the data and returns the subset of indices at the left and the right branch.
- For example, say we’re starting at the root node (so
node_indices = [0,1,2,3,4,5,6,7,8,9]), and we chose to split on feature0, which is whether or not the example has a brown cap.- The output of the function is then,
left_indices = [0,1,2,3,4,7,9](data points with brown cap) andright_indices = [5,6,8](data points without a brown cap)
- The output of the function is then,
| Brown Cap | Tapering Stalk Shape | Solitary | Edible | ||
|---|---|---|---|---|---|
| 0 |  |
1 | 1 | 1 | 1 |
| 1 |  |
1 | 0 | 1 | 1 |
| 2 |  |
1 | 0 | 0 | 0 |
| 3 |  |
1 | 0 | 0 | 0 |
| 4 |  |
1 | 1 | 1 | 1 |
| 5 |  |
0 | 1 | 1 | 0 |
| 6 |  |
0 | 0 | 0 | 0 |
| 7 |  |
1 | 0 | 1 | 1 |
| 8 |  |
0 | 1 | 0 | 1 |
| 9 |  |
1 | 0 | 0 | 0 |
Exercise 2
Please complete the split_dataset() function shown below
- For each index in
node_indices- If the value of
Xat that index for that feature is1, add the index toleft_indices - If the value of
Xat that index for that feature is0, add the index toright_indices
- If the value of
If you get stuck, you can check out the hints presented after the cell below to help you with the implementation.
# UNQ_C2
# GRADED FUNCTION: split_dataset
def split_dataset(X, node_indices, feature):
"""
Splits the data at the given node into
left and right branches
Args:
X (ndarray): Data matrix of shape(n_samples, n_features)
node_indices (list): List containing the active indices. I.e, the samples being considered at this step.
feature (int): Index of feature to split on
Returns:
left_indices (list): Indices with feature value == 1
right_indices (list): Indices with feature value == 0
"""
# You need to return the following variables correctly
left_indices = []
right_indices = []
### START CODE HERE ###
for i in node_indices:
if X[i][feature] == 1:
left_indices.append(i)
else:
right_indices.append(i)
### END CODE HERE ###
return left_indices, right_indices
Click for hints
-
Here’s how you can structure the overall implementation for this function ```python def split_dataset(X, node_indices, feature):
You need to return the following variables correctly
left_indices = [] right_indices = []
START CODE HERE
Go through the indices of examples at that node
for i in node_indices:
if # Your code here to check if the value of X at that index for the feature is 1 left_indices.append(i) else: right_indices.append(i)END CODE HERE
return left_indices, right_indices
```
<details>
<summary><font size="2" color="darkblue"><b> Click for more hints</b></font></summary>
The condition is <code> if X[i][feature] == 1:</code>.
</details>
Now, let’s check your implementation using the code blocks below. Let’s try splitting the dataset at the root node, which contains all examples at feature 0 (Brown Cap) as we’d discussed above. We’ve also provided a helper function to visualize the output of the split.
root_indices = [0, 1, 2, 3, 4, 5, 6, 7, 8, 9]
# Feel free to play around with these variables
# The dataset only has three features, so this value can be 0 (Brown Cap), 1 (Tapering Stalk Shape) or 2 (Solitary)
feature = 0
left_indices, right_indices = split_dataset(X_train, root_indices, feature)
print("Left indices: ", left_indices)
print("Right indices: ", right_indices)
# Visualize the split
generate_split_viz(root_indices, left_indices, right_indices, feature)
# UNIT TESTS
split_dataset_test(split_dataset)
Left indices: [0, 1, 2, 3, 4, 7, 9]
Right indices: [5, 6, 8]

[92m All tests passed.
Expected Output:
Left indices: [0, 1, 2, 3, 4, 7, 9]
Right indices: [5, 6, 8]
4.3 Calculate information gain
Next, you’ll write a function called information_gain that takes in the training data, the indices at a node and a feature to split on and returns the information gain from the split.
Exercise 3
Please complete the compute_information_gain() function shown below to compute
$$\text{Information Gain} = H(p_1^\text{node})- (w^{\text{left}}H(p_1^\text{left}) + w^{\text{right}}H(p_1^\text{right}))$$
where
- $H(p_1^\text{node})$ is entropy at the node
- $H(p_1^\text{left})$ and $H(p_1^\text{right})$ are the entropies at the left and the right branches resulting from the split
- $w^{\text{left}}$ and $w^{\text{right}}$ are the proportion of examples at the left and right branch, respectively
Note:
- You can use the
compute_entropy()function that you implemented above to calculate the entropy - We’ve provided some starter code that uses the
split_dataset()function you implemented above to split the dataset
If you get stuck, you can check out the hints presented after the cell below to help you with the implementation.
# UNQ_C3
# GRADED FUNCTION: compute_information_gain
def compute_information_gain(X, y, node_indices, feature):
"""
Compute the information of splitting the node on a given feature
Args:
X (ndarray): Data matrix of shape(n_samples, n_features)
y (array like): list or ndarray with n_samples containing the target variable
node_indices (ndarray): List containing the active indices. I.e, the samples being considered in this step.
Returns:
cost (float): Cost computed
"""
# Split dataset
left_indices, right_indices = split_dataset(X, node_indices, feature)
# Some useful variables
X_node, y_node = X[node_indices], y[node_indices]
X_left, y_left = X[left_indices], y[left_indices]
X_right, y_right = X[right_indices], y[right_indices]
# You need to return the following variables correctly
information_gain = 0
### START CODE HERE ###
information_gain = compute_entropy(y_node) - ((len(y_left)/len(y_node)) * compute_entropy(y_left) + (len(y_right)/len(y_node)) * compute_entropy(y_right))
### END CODE HERE ###
return information_gain
Click for hints
-
Here’s how you can structure the overall implementation for this function ```python def compute_information_gain(X, y, node_indices, feature):
Split dataset
left_indices, right_indices = split_dataset(X, node_indices, feature)
Some useful variables
X_node, y_node = X[node_indices], y[node_indices] X_left, y_left = X[left_indices], y[left_indices] X_right, y_right = X[right_indices], y[right_indices]
You need to return the following variables correctly
information_gain = 0
START CODE HERE
Your code here to compute the entropy at the node using compute_entropy()
node_entropy =
Your code here to compute the entropy at the left branch
left_entropy =
Your code here to compute the entropy at the right branch
right_entropy =
Your code here to compute the proportion of examples at the left branch
w_left =
Your code here to compute the proportion of examples at the right branch
w_right =
Your code here to compute weighted entropy from the split using
w_left, w_right, left_entropy and right_entropy
weighted_entropy =
Your code here to compute the information gain as the entropy at the node
minus the weighted entropy
information_gain =
END CODE HERE
return information_gain ``` If you’re still stuck, check out the hints below.
<details>
<summary><font size="2" color="darkblue"><b> Hint to calculate the entropies</b></font></summary>
<code>node_entropy = compute_entropy(y_node)</code><br>
<code>left_entropy = compute_entropy(y_left)</code><br>
<code>right_entropy = compute_entropy(y_right)</code>
</details>
<details>
<summary><font size="2" color="darkblue"><b>Hint to calculate w_left and w_right</b></font></summary>
<code>w_left = len(X_left) / len(X_node)</code><br>
<code>w_right = len(X_right) / len(X_node)</code>
</details>
<details>
<summary><font size="2" color="darkblue"><b>Hint to calculate weighted_entropy</b></font></summary>
<code>weighted_entropy = w_left * left_entropy + w_right * right_entropy</code>
</details>
<details>
<summary><font size="2" color="darkblue"><b>Hint to calculate information_gain</b></font></summary>
<code> information_gain = node_entropy - weighted_entropy</code>
</details>
You can now check your implementation using the cell below and calculate what the information gain would be from splitting on each of the featues
info_gain0 = compute_information_gain(X_train, y_train, root_indices, feature=0)
print("Information Gain from splitting the root on brown cap: ", info_gain0)
info_gain1 = compute_information_gain(X_train, y_train, root_indices, feature=1)
print("Information Gain from splitting the root on tapering stalk shape: ", info_gain1)
info_gain2 = compute_information_gain(X_train, y_train, root_indices, feature=2)
print("Information Gain from splitting the root on solitary: ", info_gain2)
# UNIT TESTS
compute_information_gain_test(compute_information_gain)
Information Gain from splitting the root on brown cap: 0.034851554559677034
Information Gain from splitting the root on tapering stalk shape: 0.12451124978365313
Information Gain from splitting the root on solitary: 0.2780719051126377
[92m All tests passed.
Expected Output:
Information Gain from splitting the root on brown cap: 0.034851554559677034
Information Gain from splitting the root on tapering stalk shape: 0.12451124978365313
Information Gain from splitting the root on solitary: 0.2780719051126377
Splitting on “Solitary” (feature = 2) at the root node gives the maximum information gain. Therefore, it’s the best feature to split on at the root node.
4.4 Get best split
Now let’s write a function to get the best feature to split on by computing the information gain from each feature as we did above and returning the feature that gives the maximum information gain
Exercise 4
Please complete the get_best_split() function shown below.
- The function takes in the training data, along with the indices of datapoint at that node
- The output of the function is the feature that gives the maximum information gain
- You can use the
compute_information_gain()function to iterate through the features and calculate the information for each feature If you get stuck, you can check out the hints presented after the cell below to help you with the implementation.
- You can use the
# UNQ_C4
# GRADED FUNCTION: get_best_split
def get_best_split(X, y, node_indices):
"""
Returns the optimal feature and threshold value
to split the node data
Args:
X (ndarray): Data matrix of shape(n_samples, n_features)
y (array like): list or ndarray with n_samples containing the target variable
node_indices (ndarray): List containing the active indices. I.e, the samples being considered in this step.
Returns:
best_feature (int): The index of the best feature to split
"""
# Some useful variables
num_features = X.shape[1]
# You need to return the following variables correctly
best_feature = -1
### START CODE HERE ###
max_IG = 0
for feature in range(num_features):
IG = compute_information_gain(X, y, node_indices, feature)
if IG > max_IG:
best_feature = feature
max_IG = IG
### END CODE HERE ##
return best_feature
Click for hints
- Here’s how you can structure the overall implementation for this function
```python
def get_best_split(X, y, node_indices):
# Some useful variables
num_features = X.shape[1]
# You need to return the following variables correctly
best_feature = -1
### START CODE HERE ###
max_info_gain = 0
# Iterate through all features
for feature in range(num_features):
# Your code here to compute the information gain from splitting on this feature
info_gain =
# If the information gain is larger than the max seen so far
if info_gain > max_info_gain:
# Your code here to set the max_info_gain and best_feature
max_info_gain =
best_feature =
### END CODE HERE ##
return best_feature
```
If you're still stuck, check out the hints below.
<details>
<summary><font size="2" color="darkblue"><b> Hint to calculate info_gain</b></font></summary>
<code>info_gain = compute_information_gain(X, y, node_indices, feature)</code>
</details>
<details>
<summary><font size="2" color="darkblue"><b>Hint to update the max_info_gain and best_feature</b></font></summary>
<code>max_info_gain = info_gain</code><br>
<code>best_feature = feature</code>
</details>
Now, let’s check the implementation of your function using the cell below.
best_feature = get_best_split(X_train, y_train, root_indices)
print("Best feature to split on: %d" % best_feature)
# UNIT TESTS
get_best_split_test(get_best_split)
Best feature to split on: 2
[92m All tests passed.
As we saw above, the function returns that the best feature to split on at the root node is feature 2 (“Solitary”)
5 - Building the tree
In this section, we use the functions you implemented above to generate a decision tree by successively picking the best feature to split on until we reach the stopping criteria (maximum depth is 2).
You do not need to implement anything for this part.
# Not graded
tree = []
def build_tree_recursive(X, y, node_indices, branch_name, max_depth, current_depth):
"""
Build a tree using the recursive algorithm that split the dataset into 2 subgroups at each node.
This function just prints the tree.
Args:
X (ndarray): Data matrix of shape(n_samples, n_features)
y (array like): list or ndarray with n_samples containing the target variable
node_indices (ndarray): List containing the active indices. I.e, the samples being considered in this step.
branch_name (string): Name of the branch. ['Root', 'Left', 'Right']
max_depth (int): Max depth of the resulting tree.
current_depth (int): Current depth. Parameter used during recursive call.
"""
# Maximum depth reached - stop splitting
if current_depth == max_depth:
formatting = " "*current_depth + "-"*current_depth
print(formatting, "%s leaf node with indices" % branch_name, node_indices)
return
# Otherwise, get best split and split the data
# Get the best feature and threshold at this node
best_feature = get_best_split(X, y, node_indices)
formatting = "-"*current_depth
print("%s Depth %d, %s: Split on feature: %d" % (formatting, current_depth, branch_name, best_feature))
# Split the dataset at the best feature
left_indices, right_indices = split_dataset(X, node_indices, best_feature)
tree.append((left_indices, right_indices, best_feature))
# continue splitting the left and the right child. Increment current depth
build_tree_recursive(X, y, left_indices, "Left", max_depth, current_depth+1)
build_tree_recursive(X, y, right_indices, "Right", max_depth, current_depth+1)
build_tree_recursive(X_train, y_train, root_indices, "Root", max_depth=2, current_depth=0)
generate_tree_viz(root_indices, y_train, tree)
Depth 0, Root: Split on feature: 2
- Depth 1, Left: Split on feature: 0
-- Left leaf node with indices [0, 1, 4, 7]
-- Right leaf node with indices [5]
- Depth 1, Right: Split on feature: 1
-- Left leaf node with indices [8]
-- Right leaf node with indices [2, 3, 6, 9]